Single Molecule Imaging
TEL +81-6-6879-4630
FAX +81-6-6879-4634
Overview
Various proteins in a living cell are working with corporation as bio-element nano-machine. The cell constructs the molecule system for signal transudation by these proteins, and does a complex and flexible behaviors. Our purpose is to reveal the molecular mechanism of signal transudation in an immune reaction. It is thought that the information in the cell via the proteins transmits with the participation of few molecules. Furthermore, neither the structure nor the environment in the cell is uniform. To achieve our goal, we have developed new microscopic techniques to observe directly the tempo-spatial dynamics in living cells. Here, we would like to introduce the microscopic techniques we developed.
1) Single molecular imaging by using a total internal reflection florescent microscope (TIRFM)
We have developed Single molecular imaging by using a TIRFM. The illumination by TIRF can excite the fluorescent dye nearby about 200 nm far from the glass surface by inserting the laser into the objective as do the total reflection on the boundary between the buffer and the glass. The background noise is reduced to 1/2000 by using this irradiation technique compared with a current fluorescence microscope. Therefore, a fluorescent image can be acquired with very good contrast, and we can detect the image of individual single molecule of fluorescent dyes or fluorescent proteins.
Single molecular imaging in living cell is enumerated as one of examples for bio- and/or medical applications using TIREM. We can observe the basic-process of signal transmits of the binding of ligand to the membrane, the dimerization, the complex formation, the phosphorylation, the diffusion of membrane proteins and so on. Thus, original behavior of the proteins can be observed by single molecular imaging in living cells by using TIRFM, while an electro microscope permits us to observe those in only fixed cells.
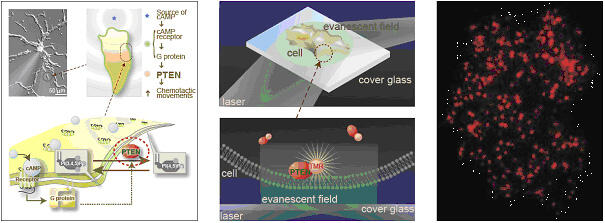
We observe PTEM, which is involved in Chemotaxis of cell, using TIRFM. The red spots are individual molecules of PTEM recruited onto the cell membrane.
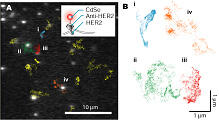
2) Nanometry of the proteins in living cells
Nanometry of the proteins in living cells We can peruse the movement of the protein by calculating the centroid of the fluorescent spot. If the fluorescent intensity is enough high, the accuracy became to be about 2 nm. We labeled HER2 (Human EGF Receptor 2), which is one of membrane proteins, with CdSe fluorescent dye (Qdot) via the antidoby, and tracked the movements of the individual fluorescent spots (Fig 2A). The membrane proteins was known to diffuse on the membrane previously, we found the diffusional movement was reflected the structures of somewhat on the membrane such as actin network, caveola or something (Fig 2B).
The nanometry in the cell enables us to observe "Life" movement of the protein that cannot be observed so far.
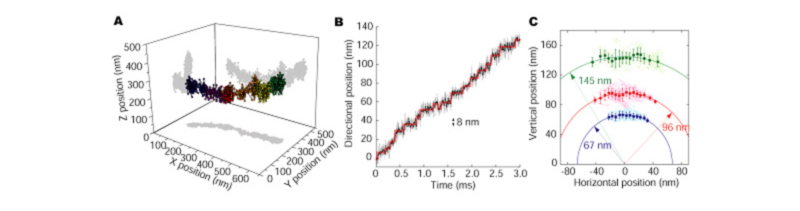
3) 3D single particle tracking
We constructed a new optics that can obtain the two distinct focal images with a CCD camera (DIO, Dual focus Imaging Optics). The principle of 3D tracking by using DIO is simple. The image of the fluorescent particle between on two focus planes is projected onto both focus images, and fluorescent intensity and the blurring of both the images correlate to the position of the vertical direction of the particle. We completed the algorithm that calculated the position of the vertical direction of the single particle by the comparing the fluorescent intensities obtained in two different focuses. XY position is determined by fitting the image with 2D Gaussian function.
We tracked the vesicle transport by dynein which is the motor protein associated with microtubule (Fig 3A). As the traveling direction of the vesicle is a coordinate axis, successive 8 nm steps were clearly detected, which indicating the small repeated structure of microtubule (Fig 3B). As the vesicle transport was viewed from the front, the vesicle was transported ahead with shaking right and left on the cylindrical column. The radius was 109 nm (Fig 3C), which consisted with the theoretical length from the center of the microtubule to the vesicle.
If the above-mentioned optics and the analysis method are used, the movement of the protein that moves in the cell can be observed as it is.
4) Inguinal skin window for chrocnic two-photo microscopic observation
Two-photon microscopy is a form of laser-scanning microscopy that uses localized nonliner optics to excite fluorescence by using near simultaneous absorption of two long wavelength (>800nm) photons. The long wavelengths used for two-photon microscopy are less damaging and penetrate more deeply into tissue than those used in other light microscopy. This distinct advantage makes it possible to perform real-time analysis of cell dynamics within their normal tissue environments in a living animal .
While two-photon microscopy to date is the only modality that can unveil cell dynamics in living animals, it is far less convenient than other imaging modalities like ultrasound and magnetic resonanse. One reason is that it is invasive, as the tissue to be observed needs to be exposed surgically by removing obstructive tissue like skin and fat. In lymphoid tissue, for example, the result is an imaging depth of a few hundreds micrometers with two-photon microscopy. Furthermore, the invasive surgical preparation makes it difficult to use one animal on different days and to follow the development of disease over a period of time.
To overcome the drawback of two-photon microscopy, we have developed an implantable observation window system for long-term observation of the mouse inguinal lymph node. To demonstrat the system's deasibility, we observed T cell migration and proliferation during homeostaic proliferation for two weeks.
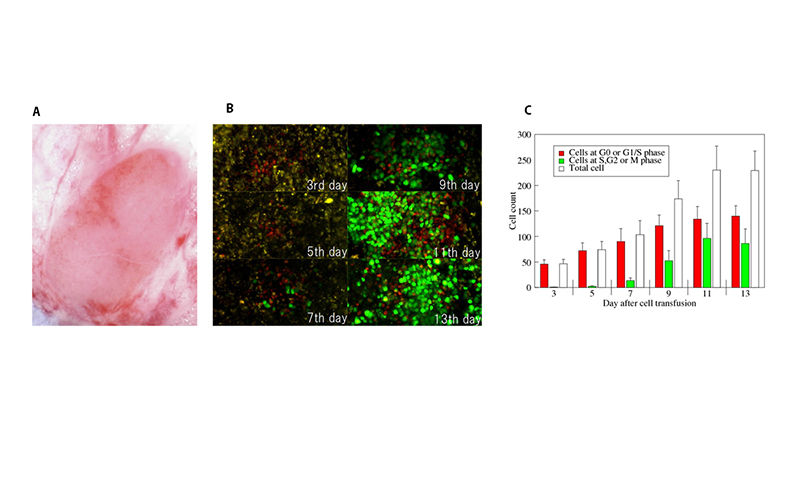
(A) A prepared inguinal lymph node in the implanted chamber.
(B) Two-photon microscopic images of T cells obtained at the same position of the same mouse from day 3 to 13.
(C) Quantified T cell proliferation from imgages.
Principal Investigator
Toshio Yanagida Professor

Research field
Single molecular imaging, Nanomerty, Optical development
Education history
| 1969 | B.A. (Engineering) Osaka University |
|---|---|
| 1971 | M.A. (Engineering) Osaka University |
| 1976 | Ph.D.(Engineering) Osaka University |
Research and career history
| 1988 | Professor, Department of Bionics, Faculty of Engineering Science, Osaka University (-2010) |
|---|---|
| 1996 | Professor, Department of Physiology and Biosignaling, Graduate School of Medicine, Osaka University (-2010) |
| 2002 | Dean, Graduate School of Frontier Bioscience, Osaka University (-2004) |
| 2002 | Professor, Graduate School of Frontier Biosciences, Osaka University (-2010) |
| 2010 | Specially Appointed Professor, Graduate School of Frontier Biosciences, Osaka University (-present) |
| 2011 | Director, Quantitative Biology Center(QBiC), RIKEN (-present) |
| 2011 | Director, Center for Information and Neural Networks(CiNet),NICT (-present) |
| 2016 | Specially Appointed Professor, Graduate School of Information Science and Technology, Osaka University (-present) |
| 2016 | Director, NEC Brain-Inspired Computing Research Alliance Laboratories (-present) |
Prize
| 1989 | 7th Osaka Science Award |
|---|---|
| 1990 | 4th Tsukahara Memorial Award (Brain Science Promotion Foundation) |
| 1992 | 1st Matsubara Lecture Award (Gordon Conference) |
| 1994 | 25th Naito Memorial Science Promotion Award (Naito Memorial Promotion Foundation) |
| 1998 | The Japan Academy Award, The Imperial Award |
| 1999 | Asahi Award (Asahi Newspaper Culture Foundation) |
| 2010 | The US Genomic Award for Outstanding Investigator in the Field of Single Molecule Fluorescence Microscopy |
| 2010 | The US Genomic Award for Outstanding Investigator in the Field of Single Molecule Biology (US Biophysical Society) |
| 2011 | Fellow of the US Biophysical Society (US Biophysical Society) |
| 2013 | Person of Cultural Merit |
| 2017 | Osaka University Distinguished Professor |
Members
-
Toshio Yanagida Professor
yanagidafbs.osaka-u.ac.jp
Achievements
Publications
-
Chiu, L.D., Ichimura, T., Sekiya, T., Machiyama, H., Watanabe, T., Fujita, H., Ozawa, T., & Fujita, K. Protein expression guided chemical profiling of living cells by the simultaneous observation of Raman scattering and anti-Stokes fluorescence emission. Sci. Rep. 7, 43569 (2017).
-
Furusawa C., Yamaguchi T. Robust and accurate discrimination of self/non-self antigen presentations by regulatory T cell suppression. PLoS One. 11(9):e0163134 (2016).
-
Ichimura, T., Chiu, L.D., Fujita, K., Machiyama, H., Yamaguchi, T., Watanabe, T.M., & Fujita, H. Non-label immune cell state prediction using Raman spectroscopy. Sci. Rep. 6, 37562 (2016).
-
Iwaki M., Wickham S., Ikezaki K., Yanagida T., Shih W. A programmable DNA origami nanospring that reveals force-induced adjacent binding of myosin VI heads. Nature Comm. 12;7:13715 (2016).
-
Fujita K., Iwaki M., Yanagida T. Transcriptional bursting is intrinsically caused by interplay between RNA polymerases on DNA. Nature Comm. 7;7:13788 (2016).
-
Morikawa T.J., Fujita H., Kitamura A., Horio T., Yamamoto J., Kinjo M., Sasaki A., Machiyama H., Yoshizawa K., Ichimura T., Imada K., Nagai T., Watanabe T.M. Dependence of fluorescent protein brightness on protein concentration in solution and enhancement of it. Sci. Rep., 6: 22342 (2016)
-
Iwaki M, Iwane A., Ikezaki K., Yanagida T. Local heat activation of single myosins based on optical trapping of gold nanoparticles. Nano Letters, 15, 2456-2461 (2015)
-
Machiyama H., Yamaguchi T., Sawada Y., Watanabe T.M., Fujita H. SH3 domain of c-Src governs its dynamics at focal adhesions and the cell membrane. FEBS J., 282(20) 4034-4055 (2015)
-
Ichimura T., Chiu L.D., Fujita K., Machiyama H., Kawata S., Watanabe T.M., Fujita H. Visualizing the appearance and disappearance of the attractor of differentiation using Raman spectral imaging. Sci. Rep., 5: 11358 (2015)
-
Karagiannis P., Ishii Y., Yanagida T. Molecular machines like myosin use randomness to behave predictably. Chem. Rev., 114 (6), pp 3318–3334 (2014)
-
Ichimura T., Chiu L.D., Fujita K., Kawata S., Watanabe T.M., Yanagida T., Fujita H. Visualizing cell state transition using Raman spectroscopy. PLoS One, 9(1) e84478 (2014)
-
Ikezaki K., Komori T., Yanagida T. Spontaneous detachment of the leading head contributes to myosin VI backward steps. PLoS One., 8(3):e58912 (2013)
-
Fujita K., Iwaki M., Iwane A. H., Marcucci L., Yanagida T. Switching of myosin-V motion between the lever-arm swing and Brownian search-and-catch. Nat. Commun., 17;3:956 (2012)
-
Morimatsu M., Togashi Y., Nishikawa S., Sugawa M., Iwane A., Yanagida T. Spontaneous structural changes in actin regulate G-F transformation. PLoS One, 7(11) e45864 (2012)
-
Ikezaki K., Komori T., Sugawa M., Arai Y., Nishikawa S., Iwane A. H., Yanagida T. Simultaneous observation of the lever arm and head explains myosin VI dual function. Small, 8(19):3035-40 (2012)
-
Ohmachi M., Komori Y., Iwane A.H., Fujii F., Jin T., Yanagida T. Fluorescence microscopy for simultaneous observation of 3D orientation and movement and its application to quantum rod-tagged myosin V. Proc Natl Acad Sci USA, 109(14):4294-8 (2012)
-
Matsuoka S., Miyanaga Y., Yanagida T., Ueda M. Visualizing single molecules in living Dictyostelium cells using total internal reflection fluorescent microscopy (TIRFM). Cold Spring Harb Protoc. 2012(3):349-51 (2012)
-
Watanabe T.M., Tsukasaki Y., Fujita H., Ichimura T., Saitoh T., Akira S., Yanagida T. Distinct modulated pupil function system for real-time imaging of living cells. PLoS One., 7(9):e44028 (2012)
-
Nishikawa S., Arimoto I., Ikezaki K., Sugawa M., Ueno H., Komori T., Iwane A.H., Yanagida T. Switch between large hand-over-hand and small inchworm-like steps in myosin VI. Cell, 142, 879-88 (2010)
-
Fujii T., Iwane A.H, Yanagida T., Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature, 467, 724-8 (2010)他300篇
Special mention
[Domestic]
President, The Biophysical Society of Japan (2002-2003)
Project Leader, Yanagida Biomotron Project, ERATO, JST (1992-1997)
Project Leader, Single Molecule Process Project, ICORP, JST (1998-2002)
Research Director, Formation of Soft Nano-Machines, CREST, JST (2002-2008)
Research Supervisor, Novel Measuring and Analytical Technology Contributions to the Elucidation and Application of Life Phenomena, CREST, JST (2004-2012)
Program Director, HPCI Program for Computational Life Sciences, RIKEN(2011-2016)
[Overseas]
Vice-Chair of the Council of Scientists, The International Human Frontier Science Program Organization (2004-2006)
Member of Council, Biophysical Society (2006-2010)
